quantitative biology + synthetic biology + systems biology
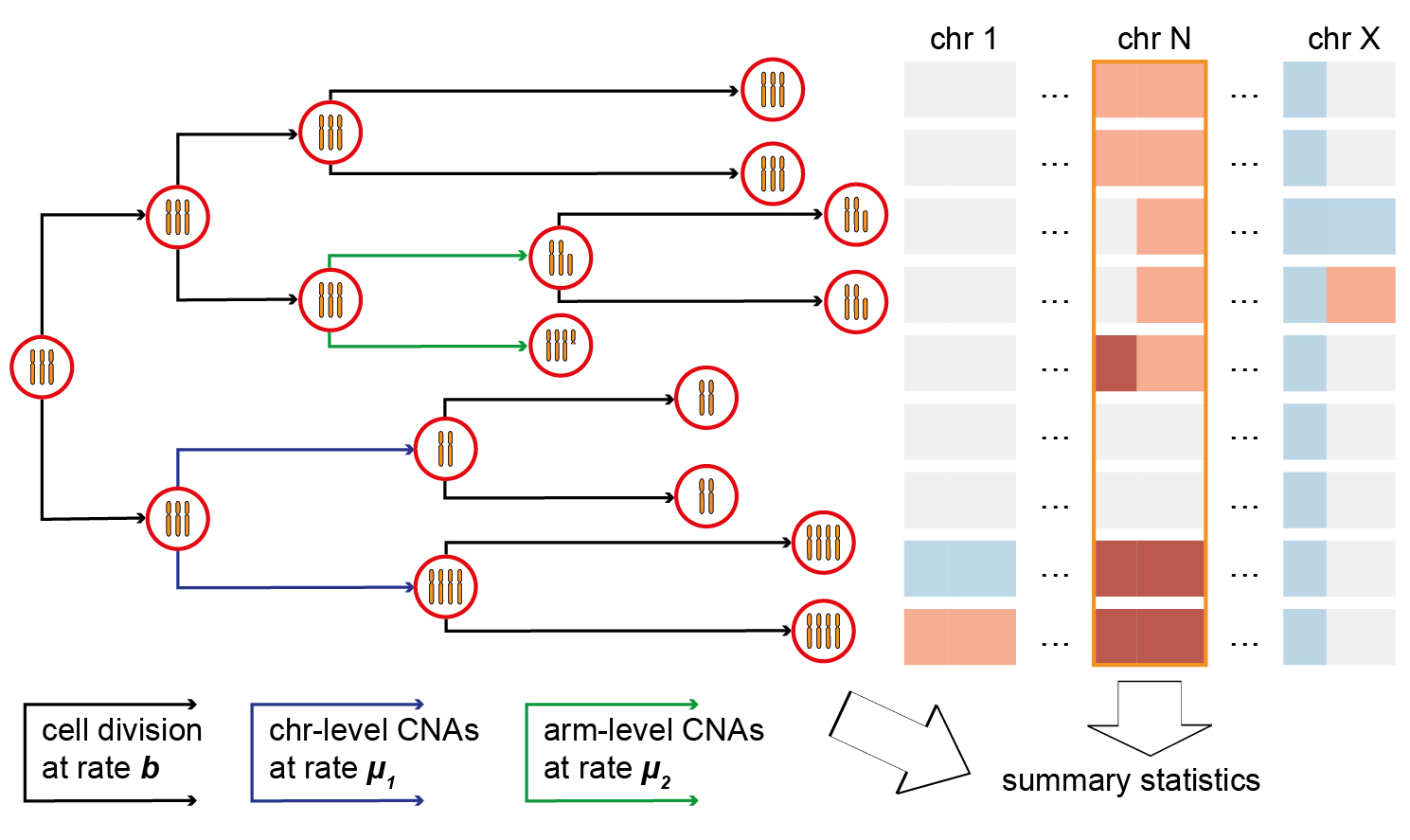
Mutational processes
Microbiome engineering
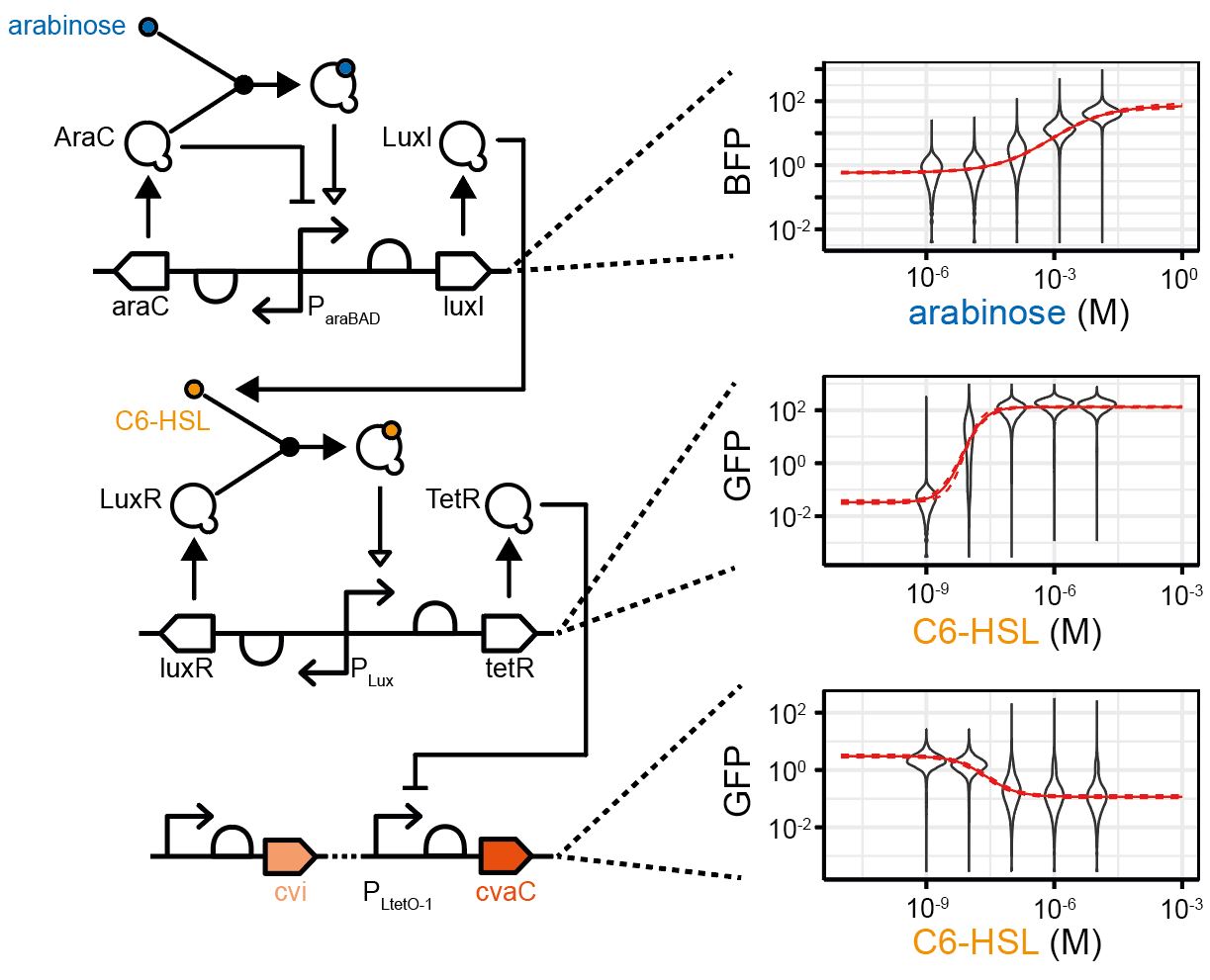
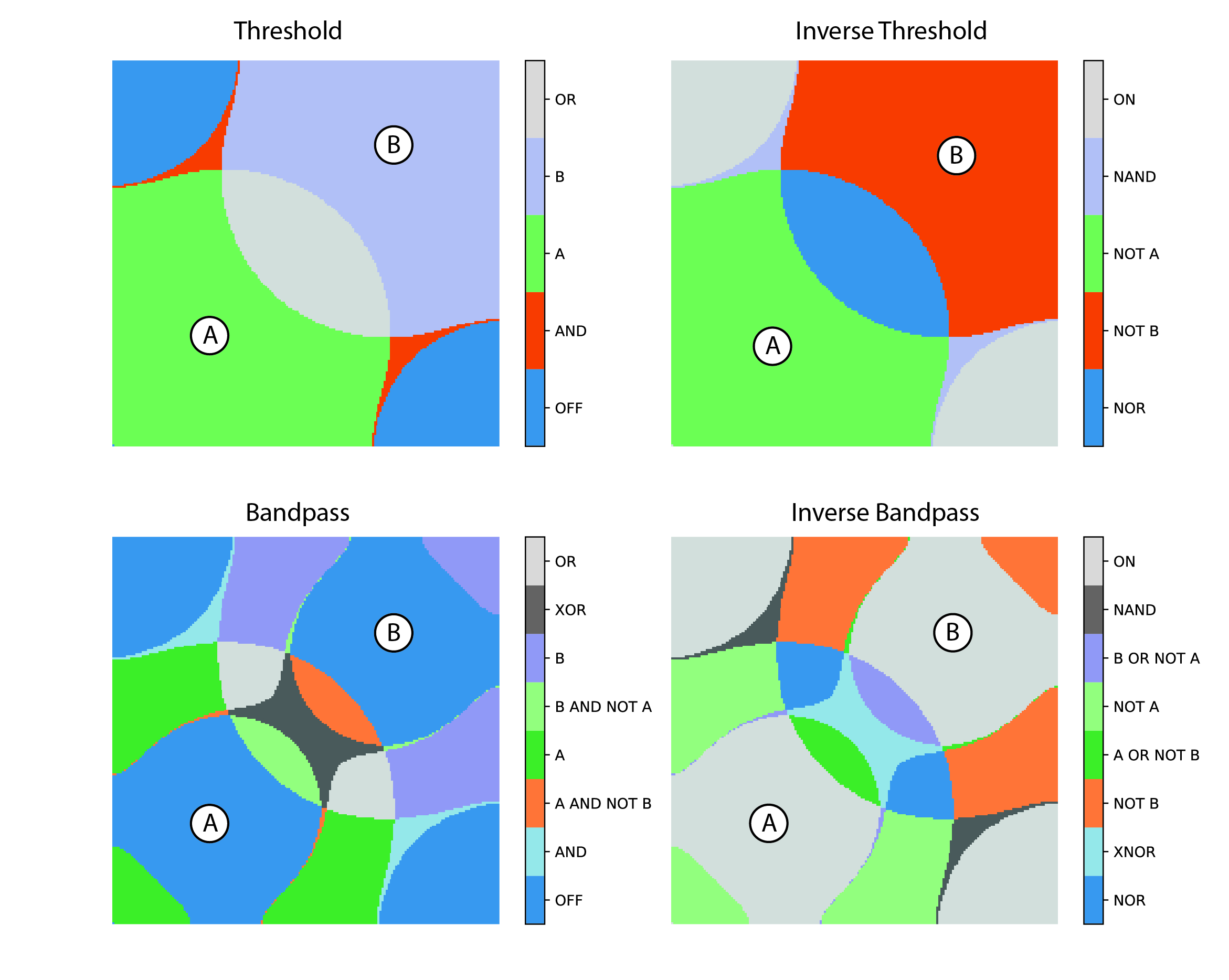
Biological computation
Mutational processes
We focus on the mechanistic modelling of genetic and sequence data to try to infer the biological processes that shape genomes. We have developed evolutionary models that can be fit to cancer patient sequencing data to infer important parameters such as mutation rates and selection coefficients. We are currently working on modelling structural variation and chromosome instability to try to understand how these important processes contribute to intra-tumour heterogeneity.
Microbiome engineering
The human intestine and the wide range of prokaryotic and eukaryotic organisms it supports form a mutualistic host-microbe symbiotic system crucial for many processes including the breakdown of plant polysaccharides and microbial fermentation. Variation and disruption of this natural ecosystem is linked to a diverse array of disorders including infectious disease, autoimmunity, obesity and cancer. We are engineering probiotic strains for therapeutic and sensing applications, plus understanding how to engineer microbial consortia.
Gene regulatory networks
Using model space exploration from Bayesian statistics we have developed and parameterised a number of models of morphogenesis in collaboration with developmental biologists. We have also used the same methods to examine the robustness of simple gene networks in the context of synthetic biology, including the dynamics of multi-functional networks.
Biological computation
Biological organisms comprise complex information processing systems and computation is present at every level, from protein and DNA nanomachines, through cellular gene and signalling networks to tissues, brains and ecosystems. Thus, computation can be seen to be a fundamental and unifying theme of biology. This computation is performed in a highly parallel and robust fashion, using concurrent, multi-scale hybrid digital and analogue processing, often utilising components that are unreliable and noisy, and at orders of magnitude lower power requirements than traditional, “silicon” computing. Understanding how biological computing is performed thus has enormous potential for driving forward biology and engineering, enabling a more sustainable future in an increasingly computation-dependent world.