(3, 100, 5)
[28.88248428 46.93130458 16.82105576 38.67933238 44.87209099]
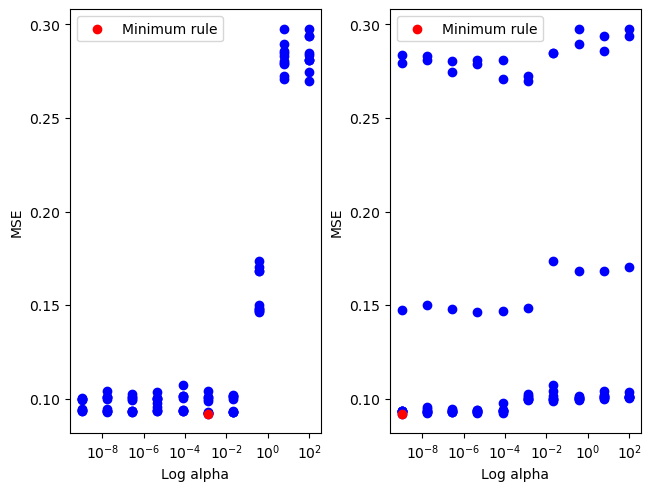
minimum found: a/error: [1.29154967e-03 1.00000000e-09] 0.09196601176191062
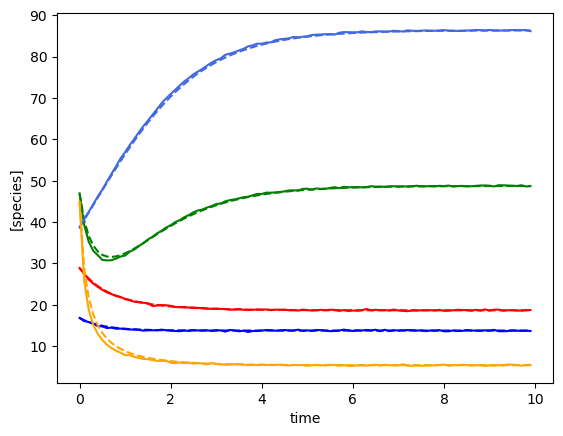
Using the following parameters for gLV simulation: {'num_species': 5, 'mu': [1.3270918724624219, 0.6434593659409151, 1.878893955402427, 0.8597855899687001, 0.8915024867127366], 'M': [[-0.053860757051265626, 0.0017940302826897423, -0.02351140030037711, -0.0010925577901253981, 0.001766638817205914], [0.0, -0.08985050006840202, -0.007911392517321557, 0.0442363991759798, 0.004590307004271697], [-0.0021297621140682647, 0.0006375198319532151, -0.13432230162469302, -0.0003385552274904023, 0.0018377092669786441], [0.0, -0.00010594551152418524, -0.0009724471160910383, -0.009743895694186036, 0.0], [0.0, 0.007257662703460285, -0.019106700711422665, -0.0022878156013554983, -0.14486771497608295]], 'epsilon': array([], shape=(5, 0), dtype=float64)}
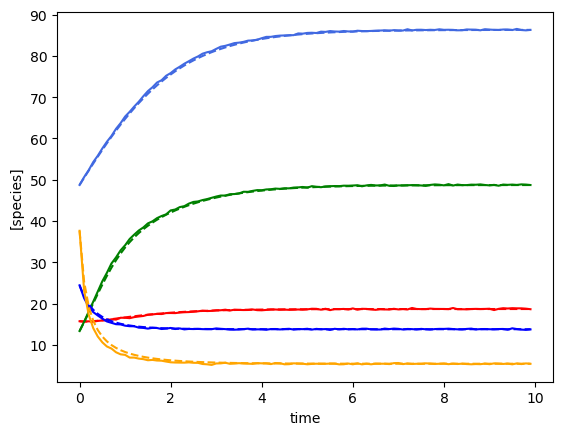
Using the following parameters for gLV simulation: {'num_species': 5, 'mu': [1.3270918724624219, 0.6434593659409151, 1.878893955402427, 0.8597855899687001, 0.8915024867127366], 'M': [[-0.053860757051265626, 0.0017940302826897423, -0.02351140030037711, -0.0010925577901253981, 0.001766638817205914], [0.0, -0.08985050006840202, -0.007911392517321557, 0.0442363991759798, 0.004590307004271697], [-0.0021297621140682647, 0.0006375198319532151, -0.13432230162469302, -0.0003385552274904023, 0.0018377092669786441], [0.0, -0.00010594551152418524, -0.0009724471160910383, -0.009743895694186036, 0.0], [0.0, 0.007257662703460285, -0.019106700711422665, -0.0022878156013554983, -0.14486771497608295]], 'epsilon': array([], shape=(5, 0), dtype=float64)}
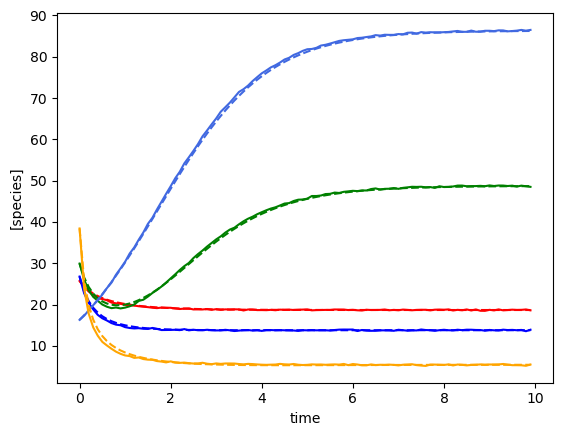
Using the following parameters for gLV simulation: {'num_species': 5, 'mu': [1.3270918724624219, 0.6434593659409151, 1.878893955402427, 0.8597855899687001, 0.8915024867127366], 'M': [[-0.053860757051265626, 0.0017940302826897423, -0.02351140030037711, -0.0010925577901253981, 0.001766638817205914], [0.0, -0.08985050006840202, -0.007911392517321557, 0.0442363991759798, 0.004590307004271697], [-0.0021297621140682647, 0.0006375198319532151, -0.13432230162469302, -0.0003385552274904023, 0.0018377092669786441], [0.0, -0.00010594551152418524, -0.0009724471160910383, -0.009743895694186036, 0.0], [0.0, 0.007257662703460285, -0.019106700711422665, -0.0022878156013554983, -0.14486771497608295]], 'epsilon': array([], shape=(5, 0), dtype=float64)}
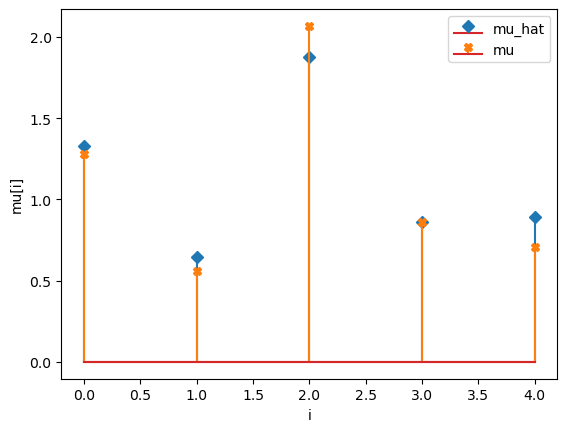
mu_hat/mu:
[1.32709187 0.64345937 1.87889396 0.85978559 0.89150249]
[1.27853844 0.55683415 2.06752757 0.86387608 0.70448068]
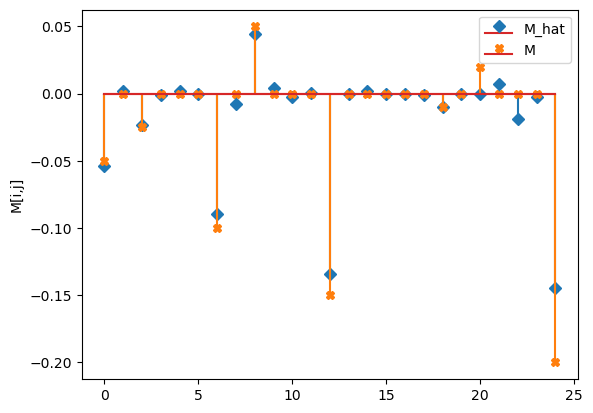
M_hat/M:
[[-0.05 0. -0.02 -0. 0. ]
[ 0. -0.09 -0.01 0.04 0. ]
[-0. 0. -0.13 -0. 0. ]
[ 0. -0. -0. -0.01 0. ]
[ 0. 0.01 -0.02 -0. -0.14]]
[[-0.05 0. -0.025 0. 0. ]
[ 0. -0.1 0. 0.05 0. ]
[ 0. 0. -0.15 0. 0. ]
[ 0. 0. 0. -0.01 0. ]
[ 0.02 0. 0. 0. -0.2 ]]